Comparison between Chemometrics and Machine Learning for the Prediction of Macronutrients in Cheese Using Imaging Spectroscopy
Accepted for poster presentation at the 38th EFFoST International Conference 2024, and submitted to Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. · Sep 11, 2024
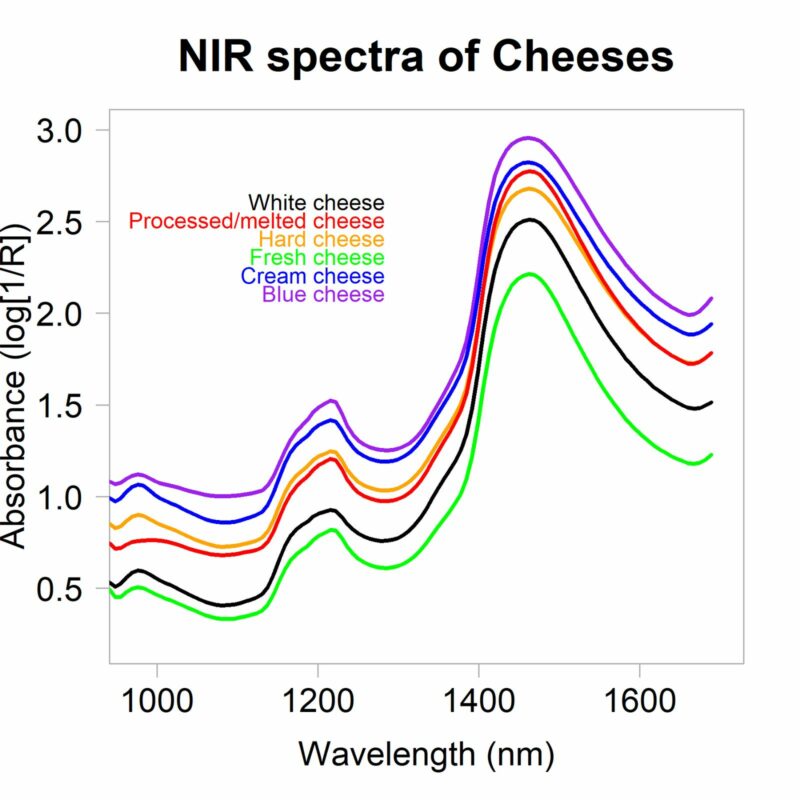
The goal of this study was to compare machine learning techniques with traditional chemometrics to predict fat and protein percentages in 73 Dutch cheeses. A comprehensive machine learning pipeline was established using multilayer perceptrons (MLP), where each macronutrient model was trained separately. Variable selection played a key role, as preprocessing methods like Standard Normal Variate (SNV), first and second derivatives, and Extended Multiplicative Scatter Correction (EMSC) were tested.
In this context, feature selection was achieved through methods like CovSel and UVE-PLS, which enabled the extraction of key wavelengths (e.g., 941.1 nm, 976.19 nm, and 1165.95 nm) crucial for predicting protein and fat content. Although the ML models could select features implicitly, the lack of interpretability underscored the “black-box” nature of such models. Chemometric approaches, on the other hand, provided transparency in the selected variables. This work emphasized the importance of integrating feature selection methods to balance performance and interpretability in models, an area where ML still faces challenges.
When comparing ML and Chemometrics, several factors were considered, including model complexity, ease of interpretation, and computational efficiency. MLP models demonstrated high complexity with multiple layers and non-linear activation functions, whereas Partial Least Squares (PLS) models in chemometrics were simpler, focusing on linear relationships. While MLPs could capture more complex patterns, they were computationally intensive and difficult to interpret. Conversely, PLS models offered greater interpretability, which is vital in food quality analysis. This balance between the intricate power of ML and the simplicity of Chemometrics underscored the need for hybrid methods that combine the strengths of both approaches.The goal of this study was to compare machine learning techniques with traditional chemometrics to predict fat and protein percentages in 73 Dutch cheeses. A comprehensive machine learning pipeline was established using multilayer perceptrons (MLP), where each macronutrient model was trained separately. Variable selection played a key role, as preprocessing methods like Standard Normal Variate (SNV), first and second derivatives, and Extended Multiplicative Scatter Correction (EMSC) were tested. In this context, feature selection was achieved through methods like CovSel and UVE-PLS, which enabled the extraction of key wavelengths (e.g., 941.1 nm, 976.19 nm, and 1165.95 nm) crucial for predicting protein and fat content. Although the ML models could select features implicitly, the lack of interpretability underscored the “black-box” nature of such models. Chemometric approaches, on the other hand, provided transparency in the selected variables. This work emphasized the importance of integrating feature selection methods to balance performance and interpretability in models, an area where ML still faces challenges. When comparing ML and Chemometrics, several factors were considered, including model complexity, ease of interpretation, and computational efficiency. MLP models demonstrated high complexity with multiple layers and non-linear activation functions, whereas Partial Least Squares (PLS) models in chemometrics were simpler, focusing on linear relationships. While MLPs could capture more complex patterns, they were computationally intensive and difficult to interpret. Conversely, PLS models offered greater interpretability, which is vital in food quality analysis. This balance between the intricate power of ML and the simplicity of Chemometrics underscored the need for hybrid methods that combine the strengths of both approaches.
Mercedes Bertotto1,3, Esther Kok1,3, Meeke Ummels1, Hajo Rijgersberg1, Guido Camps1, Edith Feskens1, Rosalba Calvini2
1. Wageningen University & Research P.O. Box 123, 6700 AB Wageningen
2. Department of Life Sciences, University of Modena and Reggio Emilia, Pad. Besta, Via Amendola, 2, 42122, Reggio Emilia, Italy
3. Shared first co-authorship
